
The 2022 Nobel Prize in Physiology or Medicine was awarded on October 3 to geneticist Svante Pääbo for his groundbreaking discoveries about human evolution and the genomes of extinct hominins.
Dr. Pääbo is director of the Max Planck Institute for Evolutionary Anthropology in Leipzig, Germany. His decades of research made it possible to extract and sequence a complete Neanderthal genome from bone fragments that were more than 40,000 years old, a task described by the Nobel Committee as “seemingly impossible.” Pääbo also made the sensational discovery of a previously unknown hominin group, the Denisovans. Importantly, Pääbo found that gene transfer had occurred from both of these extinct hominin groups to Homo sapiens following the migration out of Africa. This ancient flow of genes to present-day humans has physiological relevance today, for example affecting how our immune system reacts to infections.
Pääbo’s foundational research gave rise to an entirely new scientific discipline; paleogenomics. By revealing genetic differences that distinguish all living humans from extinct hominins, his discoveries provide a method for exploring what makes us uniquely human.
“Paleogenomics has changed the study of human origins, because it allows us to look directly at the genome to infer relationships and assess population history,” said Anne Stone, Regents’ Professor in the School of Human Evolution and Social Change at Arizona State University and member of The Leakey Foundation’s Scientific Executive Committee.
Fossils and morphology tell very important parts of the story of human evolution, but paleogenomic techniques have given scientists a new way to address important questions about humanity’s origins, including where we came from, why some species went extinct, and what makes us uniquely human.
Stone said, “The genomic analyses of archaic humans allowed Svante Pääbo and his collaborators to unexpectedly discover ‘Denisovans’ and reveal the complexity of interactions among populations in the Pleistocene. This work also allows us to examine the bits of archaic ancestry that are found in our genomes today. Svante has been at the forefront of ancient DNA research since the beginning and his lab really pushed the field forward both in terms of methodological development and the questions that he has asked. This prize is a terrific honor that is richly deserved.”
Pääbo was born in Stockholm in 1955 and was interested in early human history from a young age. He studied Egyptology and medicine at Uppsala University in Sweden. As a Ph.D. student in immunology, he demonstrated that DNA can survive in ancient Egyptian mummies.
After his doctorate, Pääbo worked in the team of evolutionary biologist Allan Wilson at the University of California at Berkeley. From 1990, he headed his own laboratory at the Ludwig Maximilian University in Munich. In 1997, Pääbo became one of five directors at the newly founded Max Planck Institute for Evolutionary Anthropology in Leipzig, where he is still active today.
As early as the mid-1990s, Pääbo and his team were able to decipher a relatively short component of the mitochondrial DNA of a Neanderthal male. Mitochondria are tiny power plants in cells that supply them with energy and have their own DNA. This Neanderthal DNA differed considerably from the genome of modern humans. This proved that Neanderthals are not the direct ancestors of today’s humans.
Since DNA sequencing methods became much more efficient in the early 2000s, Pääbo began sequencing the entire Neanderthal genome present in the cell nucleus.

A gigantic puzzle
The process of sequencing the Neanderthal genome was extraordinarily challenging. After thousands of years, the bones of Neanderthals are so heavily colonized by bacteria and fungi that up to 99.9 percent of the DNA found in them originates from microbes. In addition, the small amounts of remaining Neanderthal DNA are only present in short fragments that have to be assembled like a gigantic puzzle. Many scientists believed that this puzzle could not be solved.
However, Pääbo’s team devised new solutions. The researchers worked under “clean room conditions” comparable to those in the computer chip industry. This enabled them to prevent the inadvertent introduction of their own DNA into the experiments. In addition, they developed more efficient extraction methods that improved the yield of Neanderthal DNA. Complex computer programs that compared the DNA fragments of ancient bones with reference genomes of chimpanzees and humans helped to reconstruct the Neanderthal genome.
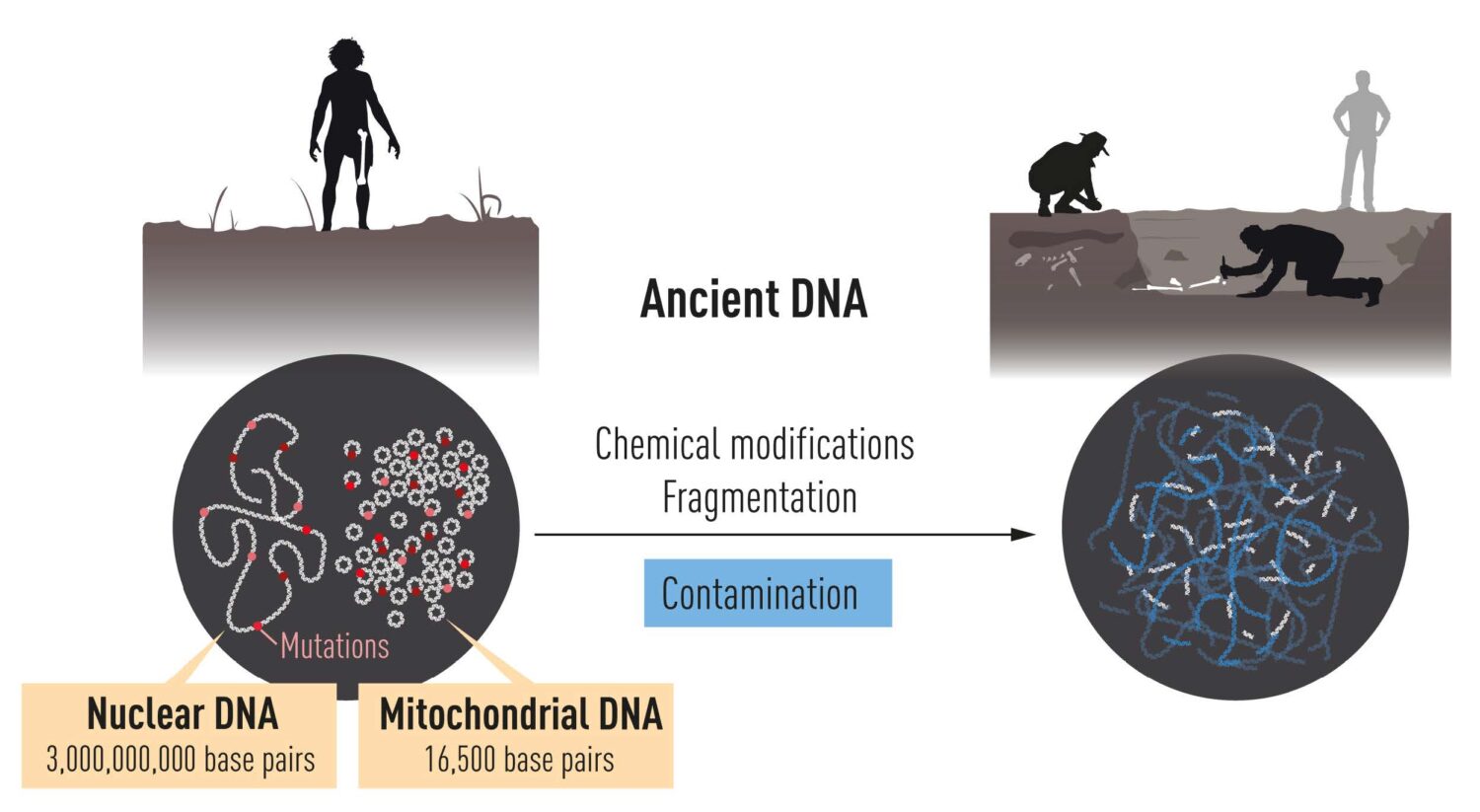
DNA is localized in two different compartments in the cell. Nuclear DNA harbors
most of the genetic information, while the much smaller mitochondrial genome is present in
thousands of copies. After death, DNA is degraded over time and ultimately only small
amounts remain.
The emergence of paleogenetics
In 2010, Svante Pääbo and his team succeeded in reconstructing the first version of the Neanderthal genome from bones tens of thousands of years old. Comparisons of the Neanderthal genome with the genomes of today’s humans showed that modern humans and Neanderthals met and produced offspring about 50,000 years ago, when modern humans left Africa and arrived in Europe and Asia.
Even today, the genome of today’s non-African people still contains about two percent Neanderthal DNA. This genetic contribution influenced human evolution: it strengthened the immune system of modern humans, for example, but still contributes to their susceptibility to several diseases.
“Neanderthals are the closest relatives of humans today,” said Svante Pääbo. “Comparisons of their genomes with those of modern humans and with those of apes enable us to determine when genetic changes occurred in our ancestors. In the future, it could also be clarified why modern humans eventually developed a complex culture and technology that enabled them to colonize almost the entire world. However, this required a more complete knowledge of the Neanderthal genome than the team had acquired in 2010.
Tracing human origins
In 2014, the team at the Max Planck Institute for Evolutionary Anthropology succeeded in deciphering the Neanderthal genome almost completely. This made a comparison with the genomes of today’s humans possible. “We have found around 30,000 positions in which the genomes of almost all modern humans differ from those of Neanderthals and great apes,” said Pääbo. “They answer what makes anatomically modern humans ‘modern’ in the genetic sense as well. Some of these genetic changes may be the key to understanding what distinguishes the cognitive abilities of today’s humans from those of now-extinct hominids.”
Prior to this, Svante Pääbo’s team had already achieved a sensation in 2012: They decoded the genome from a small bone found in the Denisova Cave in the Altai Mountains in western Siberia. The mysterious hominins were remotely related to the Neanderthals and contributed up to five percent to the genome of today’s inhabitants of Papua New Guinea, Aboriginal Australia, and other groups in Oceania.
The researchers are currently working on new methods to reconstruct DNA fragments that are even more decomposed and present in even smaller amounts. The aim is to enable research into even older DNA and genetic material from parts of the world where the survival of DNA is even rarer due to hot and humid climates.






Comments 0