Behind the Science | Journal Article

By Mareike Janiak, Leakey Foundation grantee, postdoctoral scholar, University of Calgary
At the end of 2019 a novel coronavirus emerged in Wuhan, China. This virus, named SARS-CoV-2, can lead to the disease COVID-19 in humans and has quickly spread around the world, causing tens of thousands of deaths. The pandemic is an ongoing humanitarian disaster, but concerns have also been raised about impacts of SARS-CoV-2 on non-human animals, especially our closest living relatives, the great apes and other primates.
Outbreaks of other human diseases among wild primates in the past have had fatal consequences. For example, the Ebola virus killed 5000 gorillas in 2002-2003 and yellow-fever has caused mass mortality in howler monkeys across both Central and South America. In response to SARS-Cov-2, the IUCN has released a statement warning of the potential risks of the disease to non-human primates. If they were susceptible to COVID-19, this could be especially devastating to endangered species, whose numbers have already been decimated for other reasons, such as habitat loss. However, the actual risks to non-human primates were unknown.

Without evidence of susceptibility, it is difficult to fully assess risk and more difficult to implement protective measures widely, across zoos, research facilities, and other places where human and non-human primates come into close contact, including in the wild. Our goal was, therefore, to harness the available primate genomic data to predict how susceptible they might be to the novel coronavirus. I am providing a summary of our findings (which are available as a preprint on bioRxiv) here.
Research about how SARS-CoV-2 invades human cells is rapidly emerging and has identified the ACE2 receptor, which is expressed in the lungs and other tissues, as the primary target. Part of the reason the new virus has become so widespread and deadly, is that it is very good at latching onto this receptor in humans, which it then uses to invade our cells. If you look at images of SARS-CoV-2, you’ll notice the surface of this round virus is covered in spikes.
These spike proteins bind effectively to the ACE2 protein, and thanks to the tireless efforts of researchers around the world (including some of our co-authors) we have a pretty good idea of where on ACE2 the coronavirus spike protein attaches. These attachment sites are called binding sites and are some of the individual amino acids that form the human ACE2 protein. (A great resource to learn more about this are the blog posts of the Benhur Lee Lab).
Human ACE2 proteins have a combination of amino acids that SARS-CoV-2 can attach to very effectively. But other animals do not – the virus cannot bind to the ACE2 protein in mice, for example, so it cannot invade their cells. To assess the risk to non-human primates, we looked at the ACE2 sequence across species. We then modeled the protein interactions between ACE2 and SARS-CoV-2. This approach can help us predict how susceptible non-human primates might be. If other primates have the same amino acids at these binding sites as humans do, then the virus will probably be able to bind to them very well, and the species may be at risk for COVID-19. However, if other primates have different amino acids at these sites (like mice), then they are probably less susceptible to the virus.
The figure below summarizes what we found. It shows the regions of the ACE2 protein to which the SARS-CoV-2 spike protein attaches across 27 primate species and some other mammals. The specific amino acids that are the binding sites for the virus are marked with boxes (solid boxes for the most important sites) and the human sequence across these regions is shown at the top. We can see that apes and all African and Asian monkeys are identical to humans at all of the binding sites, so they are likely as susceptible to the new virus as humans are.
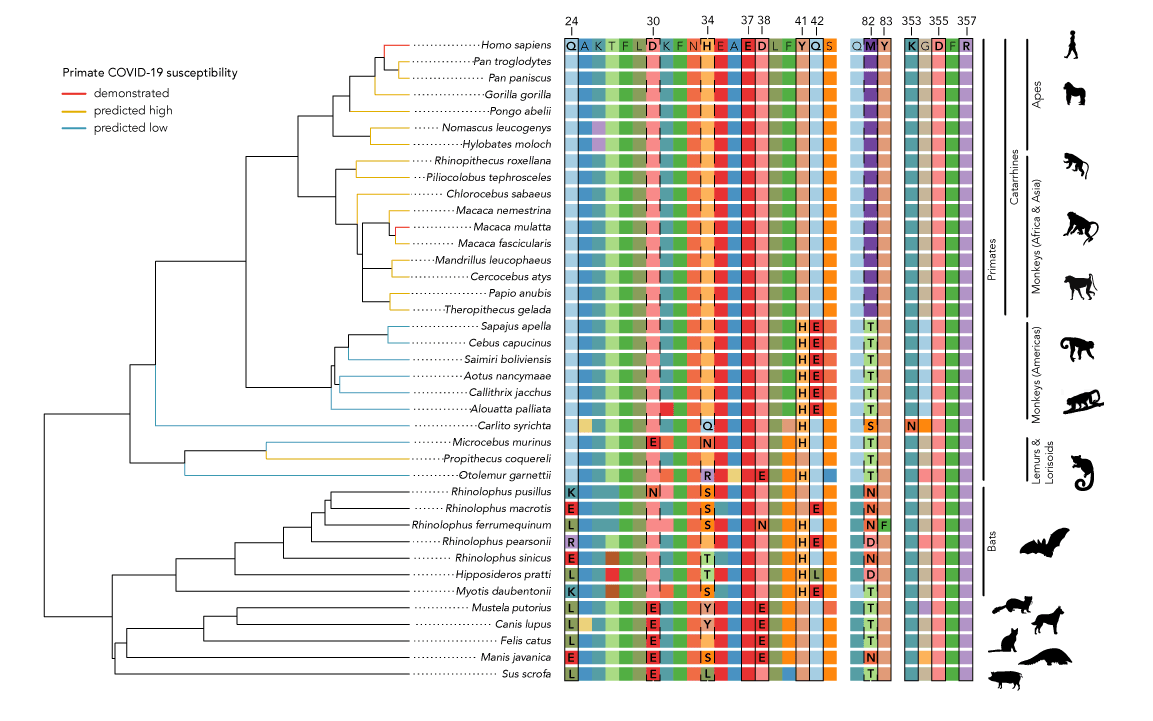
Branch lengths represent evolutionary distanceestimated from TimeTree.org. Primate species
that are able to be successfully infected with COVID-19 are indicated in red. Predicted susceptibility to COVID-19 for other primates is additionally coded by terminal branch colors.
Primates from the Americas (platyrrhines) have some differences, namely at sites 41, 42, and 82. Lemurs and galagos also show variation at various of the binding sites. Our protein interaction models suggest that the changes in the mouse lemur, galago, and platyrrhines might decrease the virus’s ability to attach to ACE2 and make them less susceptible to infection by SARS-CoV-2. Sifakas, on the other hand, likely have similar susceptibility as humans. However, we want to be very clear that these are just computational models, so the safest course of action would be to treat all primates as potentially susceptible.
Overall, our results suggest that dozens of non-human primate species, including our closest relatives, are at risk for SARS-CoV-2 infection and are vulnerable to COVID-19, which could have devastating impacts on populations of primates that are already endangered. Additional precautions may be necessary to protect captive primates and action should be taken to limit contact between wild primate populations and humans. While the human toll of the novel coronavirus has been immense and is a tragedy that requires our focus and effort, we likewise have a duty to protect our closest living relatives from its effects.
Citation:
Amanda D Melin, Mareike C Janiak, Frank Marrone, Paramjit S Arora, & James P. Higham. Comparative ACE2 variation and primate COVID-19 risk. bioRxiv 2020.04.09.034967; https://doi.org/10.1101/2020.04.09.034967